Introduction
Project PID2019-105142RB-C22 funded by
The research project Processing and crowdsourcing classification of cutaneous spindle cell neoplasm from whole slide images (Procesamiento y clasificación usando crowdsourcing de neoplasias cutáneas de células fusiformes a partir de imágenes histólogicas) is part of the project Inteligencia artificial para el diagnóstico histopatológico de neoplasias cutáneas de células fusiformes (AI4SkIN). Project PID2019-105142RB-C22 funded by MCIN/AEI/10.13039/501100011033 from 2020 to 2023.
This is a coordinated project of the Computer Vision and Behaviour Anaysis Lab (CVLAB) from the Universidad Politécnica de Valencia and the Visual Information Processing Group (VIP) from the Universidad de Granada.
The Processing and crowdsourcing classification of cutaneous spindle cell neoplasm from whole slide images research team consists of six doctors from the University of Granada, two expert pathologist from the Hospital Virgen de las Nieves de Granada, five undergraduates with extensive experience in the project theme and a doctor from the Northwestern University (Evanston, Illinois, USA).
This page will provide information on the project results and publications.
Summary
According to the World Health Organization, one in every three cancers diagnosed worldwide is a skin cancer. Unfortunately, its global incidence continues to increase. Furthermore, the workload of pathology departments is exponentially growing due to the raising number of biopsies, cancer cases, and screening programs. This creates an increasing demand to analyse biopsies which is estimated to go up to 50% in the next five years in some European regions.
This project constitutes a coordinated effort between research groups at the Polytechnic University of Valencia, the University of Granada, the Hospital Clínico Universitario of Valencia, and the Hospital Universitario San Cecilio of Granada aimed at developing a robust artificial intelligence based computer aid diagnosis system to classify, using histological images, seven types of cutaneous spindle cell neoplasms using Whole Slide Images (WSIs). Cutaneous spindle cell neoplasms are difficult to diagnose and relatively common. For example, cutaneous squamous cell carcinoma is the second most common epidermal cancer by representing 20% to 50% of skin cancers and spindle cell melanoma contributes 3% to 14% of all melanoma cases. In Spain the incidence of basal cell carcinoma and squamous cell carcinoma is 116.380 and 17.480 cases per year, respectively.
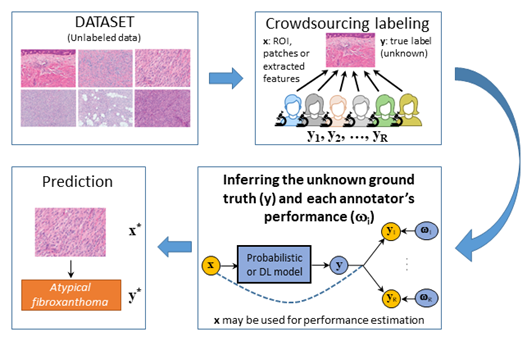
The robust system will be based on the use of images captured by different acquisition systems and stained using stains provided by different manufacturers. It will be built upon the annotation of a very large number of WSIs. This is a daunting task which cannot be carried out by a single pathologist. Different expert pathologists and pathologists in training (with different levels of expertise) will annotate overlapping subsets of WSIs. Unfortunately, the system will have to learn from a large and noisy set of annotations since to differentiate between these seven neoplasms is hard, discrepancies among experts are high, and diagnoses by less experienced pathologists are prone to errors. In other words, the system will have to be created using crowdsourcing techniques.
As a whole, the project will jointly investigate: how to efficiently pre-process and standardize histological WSI, how to extract their most significant features from automatically selected ROIs and how to find the best supervised and semi-supervised model that uses these features to diagnose utilizing crowdsourcing strategies. The areas of research of the project will help to address the challenge Health, demographic change and wellbeing. These areas are of general interest to the scientific community. The project is expected to have social and economic impact at different levels. The hospitals in the proposal will benefit from an AI solution which will help deliver accurate and fast diagnosis. Furthermore, the work of expert pathologists and pathologists in training will make it possible the annotation of huge datasets despite the limited available time of medical specialists. This will result in reliable systems which will help enhance the training of the pathologists in training. The system could be installed or used remotely by other Spanish hospitals in the future. This would open the door to a new form of collaboration between pathology departments at national level. In summary, the project represents a coordinated effort to solve interesting and challenging problems of social, medical, and scientific interest.
Objectives
The goal of the project AI4SKIN is to develop a robust artificial intelligence system to classify the following seven types of cutaneous spindle cell neoplasms: leiomyoma, dermatofibroma, atypical fibroxanthoma, leiomyosarcoma, dermatofibrosarcoma protuberans, squamous cell carcinoma and spindle cell melanoma using Whole Slide Images. This project will jointly investigate how to efficiently pre-process and standardize histological WSI, how to extract their most significant features from automatically selected ROIs and how to find the best model that uses these features to diagnose through different crowdsourcing strategies. This will facilitate and speed up the recognition of the main cutaneous spindle cell tumours, improve the training of pathologist who are confronted for the first time with this complex group of skin tumors, as well as to serve as support for more experienced pathologists but with limited experience in dermatology.
This general objective is divided in the following specific objective:
SO2.1 To develop methods for blur elimination and color normalization in WSIs. These methods will detect blurred areas and, when possible, deblur them using deconvolution techniques. Color normalization will make the system robust to inter- and intra-hospital variations. The developed techniques will help to better automatically select regions of interest extract more discriminative features, and increase the performance of machine learning methods.
SO2.2. To develop supervised crowdsourcing methods for the identification and classification of the seven types of spindle cell neoplasms based on the dataset annotated by pathologists in training. The developed methods will utilize regions of interest, features and prior information on the pathologies to jointly estimate the underlying classifier as well as the parameters characterizing each annotator’s expertise.
SO2.3 To develop semi-supervised crowdsourcing methods for the identification and classification of the seven pathologies. Due to the large size of the dataset, WSIs will not be exhaustively marked. Semi-supervised methods that use both annotated and non-annotated regions to increase the efficacy of the crowdsourcing methods will be developed.